On April 7, 2025, the research team led by Jianzhong Su at Oujiang Laboratory, in collaboration with the team led by Weihua Chen at Huazhong University of Science and Technology, published a study titled “Integrating microbial GWAS and single cell transcriptomics reveals associations between host cell populations and the gut microbiome” in Nature Microbiology. This study developed an innovative computational framework, scBPS (single-cell bacterial polygenic score), to systematically map the interaction network between the gut microbiome and host cell populations for the first time. It revealed the molecular mechanism by which the key bacterial genus Collinsella influences host health by regulating cholesterol metabolism in central vein hepatocytes, providing a novel perspective for host-microbe interaction research.
The gut microbiome is closely linked to host health, yet the mechanisms governing its interactions with specific host cell types remain poorly understood. Although genome-wide association studies (GWAS) have identified numerous host genetic loci associated with gut microbiota, little is known about how these loci regulate microbiota through tissue or cell type. Single-cell transcriptomics (scRNA-seq) offers potential for resolving cellular heterogeneity, yet studies integrating microbial GWAS with single-cell data across organs remain unexplored.

The research team pioneered the integration of metagenomic GWAS with human multi-organ single-cell transcriptomic maps. Using scBPS, they combined GWAS data from 207 gut microbial taxa within the Netherlands Microbiome Project (7,738 individuals) with transcriptomic data from 24 organs and 253 cell types covered by the Human Single-Cell Atlas (Tabula Sapiens). systematically deciphering host-microbiota interactions. By mapping genetic associations, they generated the first “microbiota-host cell interaction map.” This approach provides an intuitive tool for unraveling the microbiome's genetic mechanisms in complex diseases.
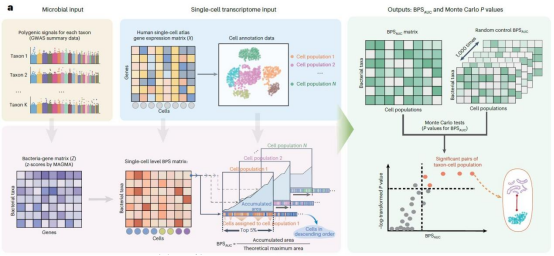
At the tissue level, modular associations between gut microbiota and host tissues were identified: - Digestive system module: Strongest associations observed in liver, pancreas, and gut tissues, particularly with Firmicutes (e.g., Lactobacillus) and Actinobacteria (e.g., Collinsella). The visual and respiratory modules: tissues like the eye and lungs showed significant associations with Actinobacteria;The muscle and immune modules: fat and immune organs exhibited weak associations with specific microbial communities.
At the cellular level, hepatic epithelial cells demonstrated the strongest association with the microbiota. Among these, the central venous hepatocyte subpopulation showed significantly stronger association with Collinsella than periportal hepatocytes. Further analysis revealed that in central vein hepatocytes with high Collinsella scores, genes related to cholesterol synthesis (e.g., HMGCR) and bile acid metabolism (e.g., CYP7A1) were significantly upregulated. Metabolic flux showed a strong positive correlation with Collinsella scores, suggesting this region is a key target for microbiota regulation of host metabolism. Using a mouse model, the research team confirmed that Collinsella influences host lipid levels by regulating cholesterol metabolism pathways in central venous hepatocytes. This discovery not only reveals the molecular basis for cross-organ metabolic regulation by gut microbiota but also provides new directions for studying the mechanisms and intervention strategies of metabolic diseases such as hypercholesterolemia.
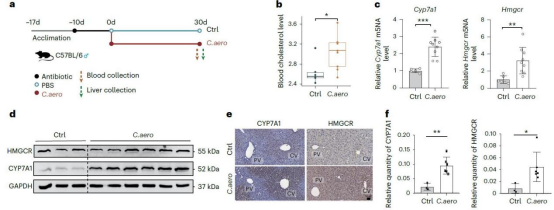
In summary, this study achieves the first three-dimensional analysis linking host genetics, microbiota, and cell types, filling a gap in the field. The development of the scBPS algorithm provides an effective tool for studying cell-specific microbial-host interactions in disease phenotypes, aiding in the precise elucidation of causal networks underlying microbiota dysbiosis in diseases and advancing the development of personalized therapies.
Manuscript URL:
https://doi.org/10.1038/s41564-025-01978-w